At 11 PM (Beijing Time) on 8th July, a new research finding was reported by the joint research team led by Prof. Rao Zihe, Distinguished Adjunct Professor at Shanghai Institute for Advanced Immunochemical Studies (SIAIS) and Professor Luke Guddat (Visiting Professor at SIAIS and Professor at University of Queensland). The related research article entitled “Structures of fungal and plant acetohydroxyacid synthases” was published via Accelerated Article Publication in Nature. This work, for the first time, showed the complete three dimensional structure of acetohydroxyacid synthase from both plants and fungi. The plant enzyme is the target for more than 50 commercial herbicides that are currently in world-wide use to protect rice, wheat and barley crops and is an emerging target for the treatment of human tuberculosis and invasive fungal pathogens such as Candida albicans. As with antibiotics, there is a growing concern over the spread of resistance to herbicides, including those whose mode of action is AHAS. It is therefore timely to enhance our understanding of the functional features of this important enzyme.
Acetohydroxyacid synthase (AHAS; E.C.2.2.1.6) is the first enzyme in the branched chain amino acid (BCAAs) biosynthesis pathway, producing L-valine, L-leucine and L-isoleucine. It consists of two subunits: (i) a catalytic subunit which contains FAD, and ThDP and (ii) a regulatory subunit which can enhance or reduce the activity of the catalytic subunits. The regulatory subunits contain the binding sites for the BCAAs, which are feedback inhibitors of the enzyme. A detailed understanding of the mechanism by which the RSUs exert their function(s) has remained a mystery dating back to the 1960s.
Our structures of Saccharomyces cerevisiae and Arabidopsis thaliana AHAS complexes demonstrate that the regulatory subunits form a core upon which the catalytic subunit dimers are attached, adopting the shape of a “Maltese Cross”. The structures show how the catalytic and regulatory subunits communicate with each other to provide a pathway for activation and for feedback inhibition by the BCAAs and how when they combine they accelerate activity. We also show that Mycobacterium tuberculosis AHAS adopts a similar highly complex structure, thus demonstrating that the overall AHAS architecture is conserved across kingdoms.
This advance represents the culmination of more than fifteen years work, overcoming many difficulties including expression of the target proteins, purification and stabilization of the complexes, and optimization of the preparation of the samples for X-ray crystallography and cryo-electron microscopy.
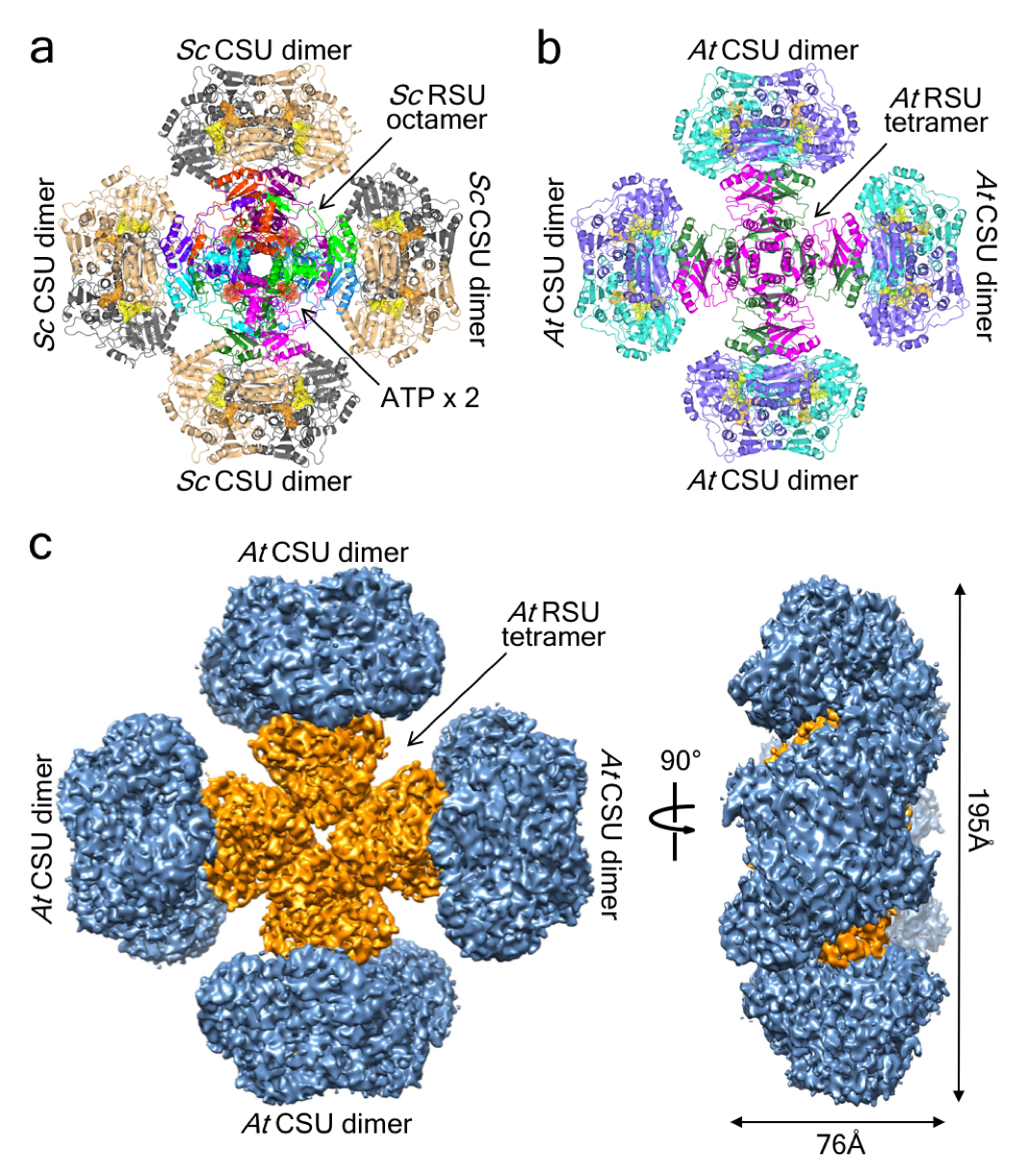
Fig. 1. The structure of the ScAHAS and AtAHAS complexes.
a. ScAHAS. Eight RSUs in different colours, each containing a single ACT domain, form a central core on which four CSU dimers are attached. Four ATP pairs (red transparent surface) are bound between RSU dimers. b. AtAHAS. Four RSUs (magenta and green), each containing two pseudo repeats of the ACT domain, form the central core. c. The cryo-EM map for AtAHAS.
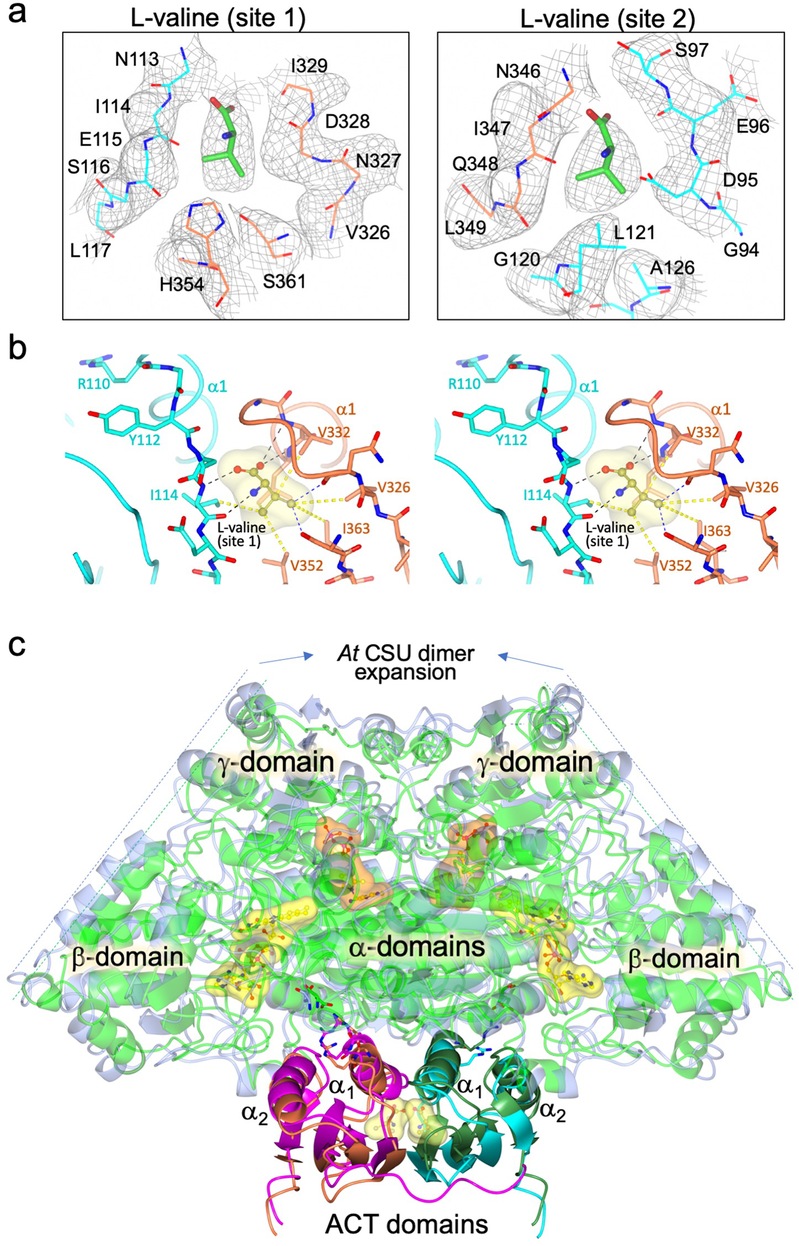
Fig. 2. Binding site interactions, and cryo-EM map for L-valine in AtRSU and consequences of L-valine binding.
a. L-Valine binding site. The cryo-EM map is overlaid. b. Interaction of L-valine in site 1 with the a1 helices of the ACT domains. c Superimposition of the free AtAHAS complex and the L-valine bound complex, representing one of the four CSU dimers and the two RSU ACT domains with which they interact. The comparison of the structures shows that the CSU dimer expands when L-valine is bound to the ACT domains of the RSUs.
This work marks a comprehensive achievement in AHAS research. The team is now taking full advantage of this structure and function study to develop next generation herbicides and new antimicrobial agents. Dr. Thierry Lonhienne and Mr. Yu Shang Low are co-first authors of the paper, Mr. Gao Yan and Professor Wang Quan from SIAS are also co-authors of this paper. The research team thanks the Bio-Electron Microscopy Platform at ShanghaiTech University and the Cryo-EM Facility Center of Southern University of Science and Technology (Shenzhen) for providing support. The authors also acknowledge the facilities and the scientific and technical assistance of the Centre for Microscopy and Microanalysis, The University of Queensland.
Link for the article:
https://www.nature.com/articles/s41586-020-2514-3



