Author:SIAIS
Date:6 June 2016
Recently, the Lerner lab has published a cover paper in the journal AngewandteChemie-International Edition (IF=11.261) titled with “Autocrine-Based Selection of Drugs that Target Ion Channels from Combinatorial Venom Peptide Libraries”. Dr. Hongkai Zhang and Mingjuan Du were co-first authors and Pro. Lerner was corresponding author.
Ion channels are important targets in drug development. A number of venom peptides have been identified to inhibit ion channels. However, venoms are difficult to isolate and analyze using traditional methods, so only a handful have been turned into drugs. The researchers of the present study demonstrated a new method by using it to identify venoms that block a certain protein on T cells--a protein implicated in multiple sclerosis, rheumatoid arthritis and other inflammatory disorders, such as ion channels Kv13.
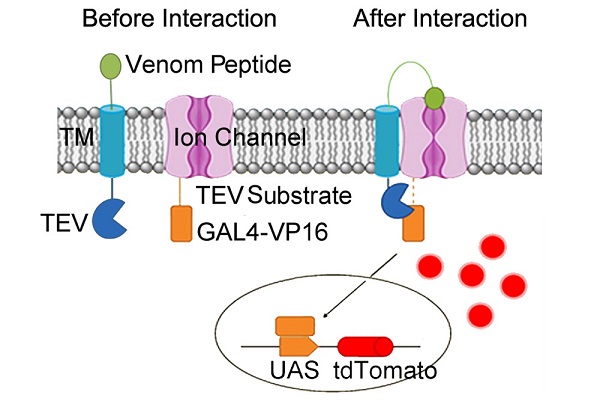
Firstly, the researchers constructed a screening system to discovery venom peptides targeting ion channels in an autocrine-based format. Toconstruct the system, the platelet derived growth factorreceptor transmembrane domain (PDGFR-TM) was used toanchor the venom(s) of interest to the plasma membrane. Thetobacco etch virus (TEV) protease was added to thecytoplasmic face of the PDGFR-TM domain. The cytoplasmic face of the voltage gated potassium channelKv1.3 was coupled to an optimized TEV substrate sequenceand the artificial transcription factor GAL4-VP16. The resultis a system with venoms bearing a protease and targetchannels bearing a substrate for the protease.When the Kv1.3channel and venom peptides interact, they bring the enzymeand its substrate into proximity, and catalytic cleavage of thesubstrate sequence occurs. The released transcription factorthen enters the nucleus and activates the expression of thefluorescent protein reporter gene. The cells expressing fluorescent protein can be sorted by FACS.
Then, to validate the screening system, the researches consulted animal toxin databases and assembled a list of 589 venoms whose protein sequences have features of interest. They then synthesized the venoms' genes and inserted them intothe system using lentivirus method. After rounds of selection, the team soon identified 27 likely Kv1.3-blocking venoms. All but two turned out to be known blockers of the ion channel. Another had been reported in the literature as a suspected potassium-channel blocker, and the last, an uncharacterized scorpion venom called CllTx1, proved in subsequent traditional-method testing--using actual venom extracted from a scorpion--to be a potent Kv1.3 blocker.
Furthermore, the team realized that their selection system could be useful not only for screening libraries of natural venoms but also for screening artificial variants or analogs of a given venom to find those with optimal pharmaceutical properties. To demonstrate, they generated about a million analogs of a long-acting protein based on ShK, a sea anemone toxin that blocks Kv1.3, and put the analogs through three rounds of selection to find the best one. The resulting candidate, S1-2, showed a strong effect not only for blocking Kv1.3 but also for reducing inflammation in a standard rodent model. This analog appears to be very potent against Kv1.3 and has no off-target effects on closely related ion channels.
Journal Reference:
1. Hongkai Zhang, Mingjuan Du, JiaXie, Xiao Liu, Jingying Sun, Wei Wang, Xiu Xin, Lourival D. Possani, Kyungmoo Yea, Richard A. Lerner.Autocrine-Based Selection of Drugs that Target Ion Channels from Combinatorial Venom Peptide Libraries. AngewandteChemie International Edition, 2016; DOI: 10.1002/anie.201603052