
Research in this lab is biological question driven. This means that we would not be limited by technique hurdles, instead we use an array of different methods, such as x-ray crystallography, mass spectrometry, cryo-EM, SAXS, high-throughput screening, deep sequencing, in vitro & in vivo evaluation etc., to investigate biological questions with clinical significance. Training in this lab entails scientific curiosity and persistence, encourages critical thinking and cooperation, while it also offers a free climate of interdisciplinary research and a chance to master difference techniques.
We study the molecular basis of a range of high-risk infectious diseases and genetic diseases, so as to identify novel therapeutic vulnerabilities in organisms. On top of these basic research, we seek to translate gained knowledge into the development of preventive opportunities like vaccine immunogens, and therapeutic interventions such as gene therapies.

Our representative research achievements are briefed below:
1. Development and application of gene editing tools: In recent years, nuclear and mitochondrial base editors have been developed by coupling the “locators” like CRISPR/Cas9 or TALE proteins with the “effectors” like APOBEC or DddA to enable base substitutions at defined sites in nuclear and mitochondrial genomes. However, the first-generation of such editors exhibited certain levels of off-target and bystander editing, limiting their broad application in both research and clinical settings. In the past, we have leveraged our expertise in protein engineering to contribute in the development of several high-precision nuclear genome editing tools, including a transformer base editor (tBE) with no detectable off-target effect (Nat Cell Biol, 2021). More recently, we captured the structural snapshots of DdCBE, the mitochondrial base editor, targeting two native mitochondrial gene loci, thus revealing the structural determinants of its editing window. Based on the mechanistic insights, we then developed a “WinPred” model to guide TALE-recognition region and spacer length design and engineered a high-precision variant of DdCBE, i.e., aDdCBE with a 2~3 nt editing window and minimal off-target mutations. Further application of the WinPred model and aDdCBE enabled near single-nucleotide editing at multiple target sites and allowed faithful modelling of Leber hereditary optic neuropathy disease-related mutations (Mol Cell, 2025) (Fig.1). Besides gene editing tools development, we are also actively exploring their application in high-risk infectious disease prevention and hereditary diseases treatment. For instance, we and our collaborators used tBE to disrupt transcription factor binding motifs in the promoter region of γ-globin in hematopoietic stem cells, thereby reactivating the silenced γ-globin expression for β-thalassemia treatment (Cell Stem Cell, 2023).
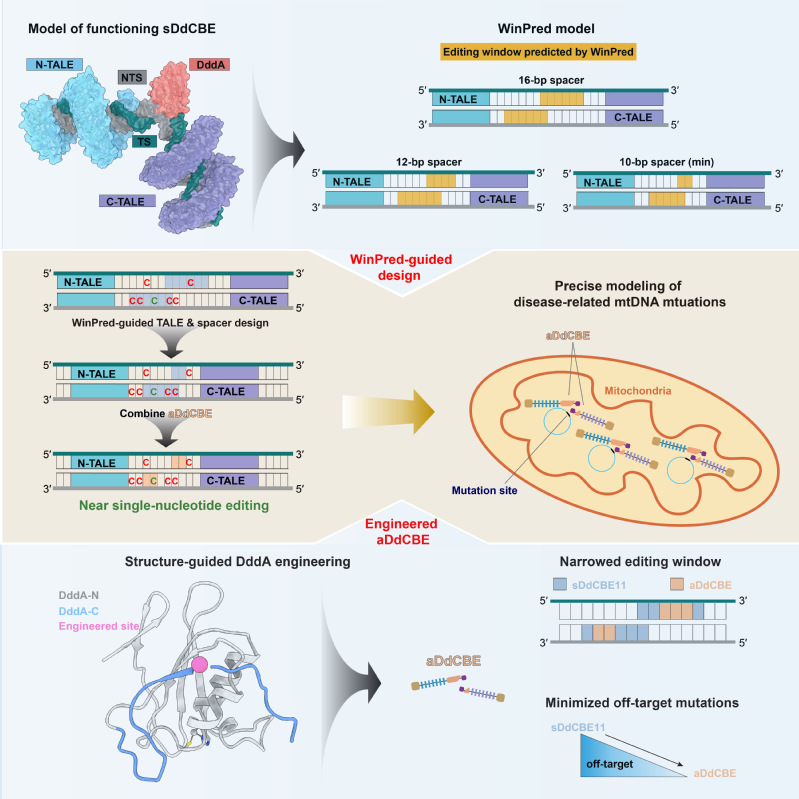
Fig.1| Structure-guided development of high-precision mitochondrial base editors
2. Vaccine targets Identification: Revealing conserved immune vulnerabilities in potential immunogens is essential for effective and broad-spectrum vaccine development. The trimeric Env protein on HIV-1 particles has long been the focus of AIDS vaccine design. However, its substantial sequence variability across HIV-1 subtypes and extensive glycosylation together impeded vaccine development for decades. To enable the application of state-of-the-art “epitope-focusing” vaccine strategy on HIV-1, we characterized the structural and immunogenic landscape of Env proteins from Asia prevalent HIV-1 subtypes, CRF01_AE and CRF07_BC. Our work identified CRF01_AE-specific features in the Env V1 region (Fig. 2A–B), uncovered their association with resistance to certain broadly neutralizing antibodies (bNAbs) (Fig. 2C–D), and revealed a novel neutralization mechanism of the first bNAb isolated from a CRF01_AE-infected individual (Fig. 2E) (Nat Commun, 2023). These findings expanded our understanding of Asia-prevalent HIV-1 subtypes and shed lights on future “epitope-focusing” HIV-1 vaccine design. Meanwhile, to better prepare for potential emergent coronaviruses, i.e., “HCoV-X”, we mapped the antigenic landscape of α-coronavirus spike proteins and revealed both conserved and divergent antigenic features across human coronaviruses, thus providing clues for future development of broadly effective coronavirus vaccines (Commun Biol, 2022).
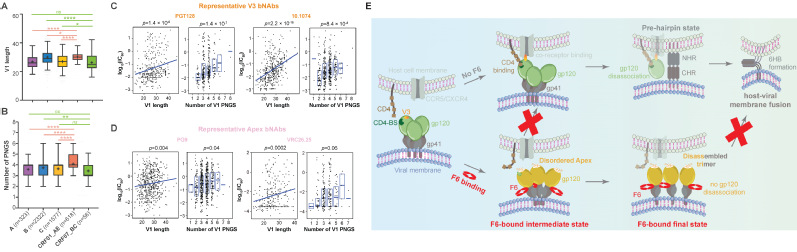
Fig.2| Structural and immunogenic understanding of Envs from CRF subtypes
3. Drug targets identification and inhibitors screening: Compared to the huge health and economic burden imposed by Enterovirus infections on society, the countermeasures have fallen short. Viral 3C protease is one of the drug development focuses of Enterovirus, yet its protease active center has frustrated previous drug discovery efforts. Using an integrative approach which combines mass spectrometry and X-ray crystallography, we demonstrated that 1) the protease activity of 3C could be modulated allosterically and 2) 3C plays a regulatory role in enteroviruses genome replication by binding to the 5’NCR of viral genome. Notably, both the allosteric site and the 5'NCR-binding site on 3C are highly conserved and locate away from the protease active center, making them attractive broad-spectrum drug targets for enterovirus infection control (PNAS, 2020). In silico screening of 143,621 natural products against the allosteric site further identified dihydromyricetin (DHM), a natural product that can broadly binds and allosterically inhibits the protease activities of 3C proteins from multiple enteroviruses. Notably, DHM shows minimal cytotoxicity and potent antiviral efficacy in different cell models, exhibiting a selective index exceeding 700. These findings establish DHM as a unique, broad-spectrum allosteric inhibitor of Enterovirus 3C proteases and underscore its potential as a promising candidate for the development of pan-enterovirus antivirals (Adv Biol, 2025).