Recently, professors Rao Zihe and Yang Haitao at SIAIS (Shanghai Institute for Advanced Immunochemical Studies) of ShanghaiTech University, together with their collaborators, have determined the high-resolution crystal structure of main protease (Mpro) from COVID-19 virus and identified drugs that may have promise against COVID-19. Invited by Nature magazine, this research was published online in Nature on April 9th 2020.
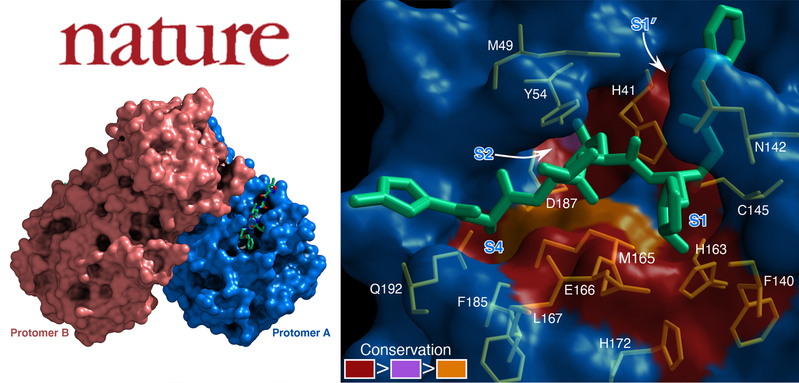
(Left) The crystal structure of COVID-19 virus Mpro in complex with N3; (Right) Surface presentation of conserved substrate-binding pockets of 12 CoV Mpros.
Professors Rao Zihe and Yang Haitao have been engaged in the research against coronavirus disease for years since SARS (Severe acute respiratory syndrome) broke out in 2003. They determined the crystal structure of main protease from SARS virus and discovered inhibitors for coronavirus. In January 2020, when the epidemic of COVID-19 broke out, professors Rao Zihe, Yang Haitao, Jiang Hualiang (professor at SIAIS and Shanghai Institute of Materia Medica), and their collaborators immediately joined in the race against the new virus as a joint research team.
Mpro, a key CoV enzyme, plays a pivotal role in mediating viral replication and transcription, making it an attractive drug target for this virus. To identify new drug leads that target the Mpro from COVID-19 virus, the team initiated a program of combined structure-assisted drug design, virtual drug screening and high-throughput screening in the research.
Here, the team identified a mechanism-based inhibitor, N3, by computer-aided drug design and subsequently determined the crystal structure of COVID-19 virus Mpro in complex with this compound on January 26th, which is the first determined public-domain 3D structure from COVID-19 virus. Next, through a combination of structure-based virtual and high-throughput screening, they assayed over 10,000 compounds including approved drugs, drug candidates in clinical trials, and other pharmacologically active compounds as inhibitors of Mpro. Among them, six of these inhibit Mpro with IC50 values ranging from 0.67 to 21.4 μM, and Ebselen also exhibited promising antiviral activity in cell-based assays.
In this study, the convergence of structure-based ab initio drug design, virtual screening and high-throughput screening proved to be an efficient strategy to find antiviral leads against COVID-19 virus. The methods presented here can greatly assist in the rapid discovery of drug leads with clinical potential in response to new emerging infectious diseases that currently lack specific drugs and vaccines.
Prior to the official release of the results, the joint team disclosed their findings in advance to the research community and the general public. On January 25th and January 26th, the team released the list of candidate drugs and the structure of COVID-19 virus main protease to the public, respectively. Till the structure was officially released on PDB (Protein Data Bank, PDB), the team had already provided data to over 300 research teams from academia and industries worldwide. The rapid release of the research data has promoted research efforts around the world and assisted clinicians, researchers and other related professionals to establish a better understanding of the new virus and develop treatments and vaccines to contain further spread.
In February 2020, the crystal structure of COVID-19 virus Mpro (3CL protease) was selected as February Molecule of the Month by PDB, and was featured in PDB news. PDB news commented, “Rapid public release of this structure of the main protease of the virus (PDB 6lu7) will enable research on this newly-recognized human pathogen”.
Read the article: https://www.nature.com/articles/s41586-020-2223-y;
DOIhttps://doi.org/10.1038/s41586-020-2223-y



