On April 10, 2020, a joint research team led by Rao Zihe, Distinguished Adjunct Professor at SIAIS (Shanghai Institute for Advanced Immunochemical Studies), ShanghaiTech University, Wang Quan, Assistant Professor at School of Life Science and Technology (SLST) and SIAIS, ShanghaiTech University and Lou Zhiyong, Research Professor at Tsinghua University, published their research findings in Science, entitled “Structure of the RNA-dependent RNA polymerase from COVID-19 Virus”. In this research, the joint team reported a 2.9 Å cryo-EM structure of the RNA-dependent RNA polymerase from COVID-19 Virus together with cofactors, which is a major target for antiviral agents, such as Favipiravir and Remdesivir.

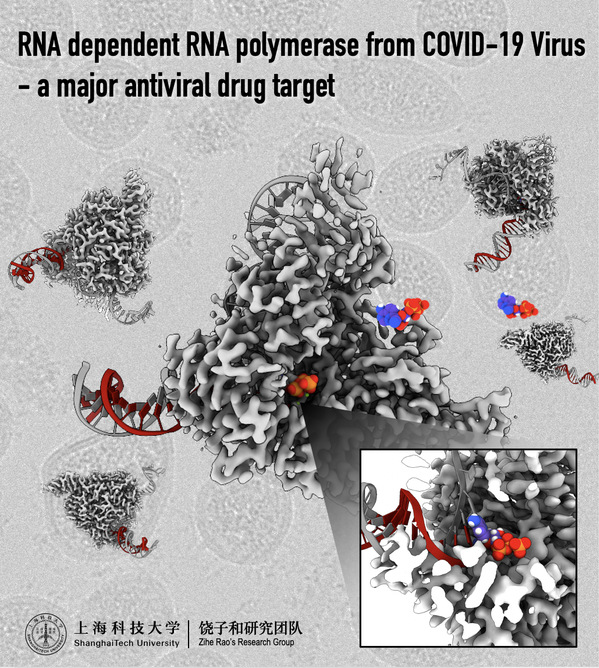
Coronavirus disease 2019 (COVID-19) virus is spreading rapidly across the world and has been announced as a “global pandemic” by the World Health Organization (WHO) on March 15, 2020. Globally, 1,439,516 confirmed cases of COVID-19, including 85,711 deaths, have been reported to WHO, as of Apr 10, 2020. The COVID-19 virus is a new member of the betacoronavirus genus and closely related to severe acute respiratory syndrome coronavirus (SARS-CoV) and Middle East respiratory syndrome coronavirus (MERS-CoV). A COVID-19 virus infection generally causes flu-like symptoms, such as fever, dry cough and fatigue. Severe cases may progress to multi-organ failure and even death. Unfortunately, no vaccine or specific medicine are available to prevent or treat COVID-19 currently.
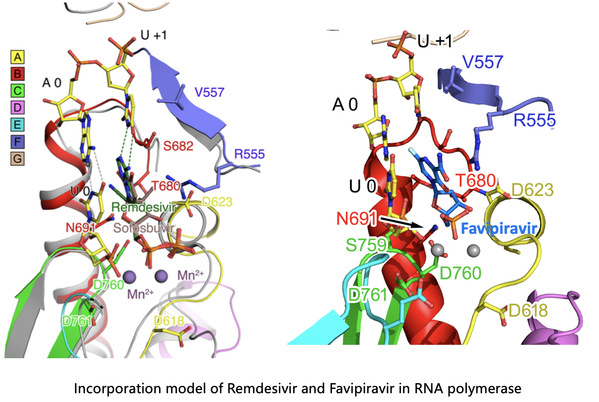
Coronaviruses employ a multi-subunit replication/transcription machinery, comprising of a set of non-structural proteins (nsp) produced as cleavage products of the ORF1a and ORF1ab viral polyproteins to facilitate virus replication and transcription. The RNA-dependent RNA polymerase (RdRp, also named nsp12), which catalyzes the synthesis of viral RNAs, is a key component of the coronaviral replication/transcription machinery and a primary target for antiviral drugs. Given the crucial function of nsp12, the characterization of its structure in complex with its cofactors nsp7 and nsp8 could provide atomic-level details to facilitate rational antiviral drug design and development. Favipiravir (an antiviral drug) and Remdesivir (an investigational antiviral compound, development code GS-5734) are considered as potential therapeutics for COVID-19 and are both undergoing clinical trials in COVID-19 patients.
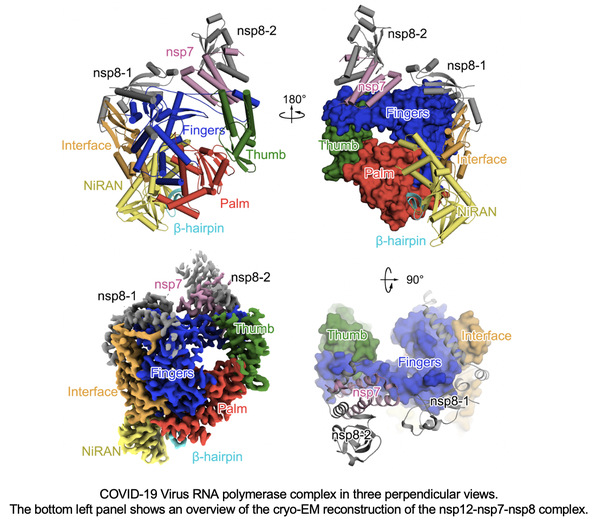
The joint research team led by Professors Rao Zihe, Wang Quan, and Lou Zhiyong, reported a cryo-EM structure of COVID-19 virus full-length nsp12 in complex with cofactors nsp7 and nsp8, at a resolution of 2.9 Å. Besides the conserved architecture of the polymerase core of the viral polymerase family and a nidovirus RdRp-associated nucleotidyltransferase (NiRAN) domain featured in coronaviral RdRp, nsp12 also possesses a newly identified β-hairpin domain at its N-terminal, where key residues for viral replication and transcription can be observed. A comparative analysis showing how Favipiravir and Remdesivir bind to this polymerase is also provided.
This structure provides insight into the central component of coronaviral replication/transcription machinery and sheds light on the design of new antiviral therapeutics targeting coronaviral RdRp.
In order to share the findings to the research community and the general public in time, the research work was at first published on bioRxiv website on March 17, 2020, entitled “Structure of RNA-dependent RNA polymerase from 2019-nCoV, a major antiviral drug target”.
To assist more researchers with their work on COVID-19, especially in the drug discovery field, the data of this cryo-EM structure has been submitted to the Protein Data Bank (PDB), and are open for download via PDB IDs 6M71 and 7BTF.
Besides, the team will also provide genes, plasmids and other experimental materials of this research to the public for free. Please contact the research team if you need any of the materials (contact person: Dr. Yang Xiuna, Research Associate Professor at SIAIS, email: yangxn@shanghaitech.edu.cn). Till now, the team has shared the cryo-EM structure data to several research institutions and companies, including Harvard Medical School, Cambridge University, University of California, Fudan University, Chinese Academy of Sciences and Chinese Academy of Medical Sciences et. al. They also has provided experimental materials to University of Queensland, University of Auckland, Cambridge University, Columbia University, Stanford University, University of California, Berkeley and other research institutions around the world.

During the COVID-19 pandemic, ShanghaiTech University has provided tremendous efforts to the joint research team. The strong support from research facilities, campus services, laboratory supplies and healthcare services, ensured a safe and supportive environment for the joint research team to successfully carry out this challenging project and attain this important achievement.
Read the article:
https://science.sciencemag.org/content/early/2020/04/09/science.abb7498



