The 3C protease has long been the focus of enterovirus antiviral development. Nevertheless, although anti-enterovirus drugs have a potentially enormous global market, the nucleophilic active site of 3C has frustrated previous drug discovery efforts. Recently, a team of scientists led by Dr. Yang Bei and Dr. Ian A. Wilson from the Laboratory of Antibody Structure at the Shanghai Institute for Advanced Immunochemical Studies (SIAIS) of ShanghaiTech University identified two novel sites of therapeutic vulnerability on 3C, both of which are distinct from its known active center. The work also demonstrates for the first time that the regulatory role of 3C in enterovirus replication is another drug-targetable function besides its known protease activity. Their work entitled “Inhibitory antibodies identify unique sites of therapeutic vulnerability in rhinovirus and other enteroviruses” was published online in Proceedings of the National Academy of Sciences (PNAS) on May 28, 2020, by authors Meng Bing, Lan Keke, Xie Jia, Richard A. Lerner, Ian A. Wilson, and Yang Bei.
As the prototype and largest genus within Picornaviridae, Enterovirus encompasses many important human pathogens such as poliovirus. Among them, Rhinovirus (HRV) alone accounts for more than 50% of common colds, and could also lead to asthma and life-threatening chronic obstructive pulmonary disease (COPD) exacerbation in young children and some adults. Moreover, poliovirus infection manifests a paralysis rate as high as 1/200, and may trigger respiratory arrest or even death in severe cases. Meanwhile, coxsackievirus and enterovirus are main causes of the hand, foot and mouth disease (HFMD) and aseptic meningitis, besides being one of the pathogenic factors of type I diabetes. Compared to the huge health and economic burden imposed by enterovirus infections on society every year, the prevention and treatment countermeasures fell short. To date, vaccines are only available for enterovirus A71 (EV71) and poliovirus. For the other viruses in Enterovirus, vaccine development is compounded by multiple serotypes. As such, a more rational strategy against enterovirus infections might be to develop broad spectrum antivirals by targeting highly conserved proteins that are indispensable for the virus life-cycle, such as the 3C protease.
3C cleaves not only the viral polyprotein into structural and non-structural viral proteins for virus progeny assembly, but also a variety of host proteins to subvert host translation, transcription and anti-viral systems. As such, 3C has been a major target for development of therapeutics against enteroviruses. However, the active site of 3C has frustrated drug discovery efforts in past decades, as most substrate analogue inhibitors contain electrophilic warheads with unfavorable pharmacokinetic properties and off-target promiscuity.
Previously, Xie, J. et al. identified two single-chain antibody fragments (scFvs), YDF and GGVV, from intracellular screening, which exhibited outstanding inhibitory effects against HRV proliferation in HeLa cells (Xie, J. et al., 2014, Chem Biol. 21:274-83). Although 3C is the target of the two scFvs, the mechanism by which they inhibit HRV infection likely differs, as YDF inhibits the protease activity of HRV 3C while GGVV does not. This finding suggested that GGVV may hinder HRV replication though inhibiting a yet unexplored function of 3C that is distinct from its protease activity, and would therefore represent a new chemotherapeutic opportunity.
In this work, Dr. Meng Bing, postdoctoral fellow in the Laboratory of Antibody Structure at SIAIS, determined the co-crystal structures of HRV 3C in complex with YDF or GGVV. As expected, the two scFvs bind to different epitopes on HRV 3C, but neither overlaps with the active site (Figure 1).
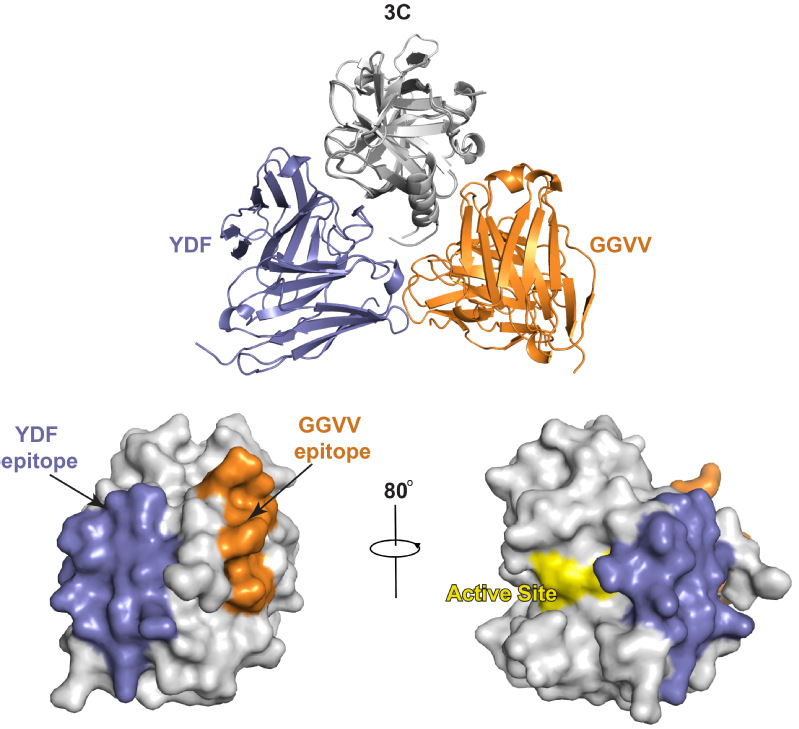
Figure 1. The epitopes of YDF and GGVV on 3C are distinct from its proteolytic active site (Top) Structural superimposition of 3C-YDF and 3C-GGVV complexes. (Bottom) YDF and GGVV bind to different surfaces on HRV 3C, both of which are distinct from the active site. The contact regions of YDF on HRV 3C are highlighted in purple; and the epitope of GGVV is highlighted in orange. The three critical residues for proteolytic activity in the active site of HRV 3C are shown as a yellow surface
Enzyme kinetics and molecular simulations of 3C then revealed that YDF inhibits the protease activity of 3C through a non-competitive, allosteric effect that is rarely seen for anti-proteolytic antibody inhibitors (Figure 2). Thus, the YDF epitope represents an alternative target site for modulating the proteolytic function of 3C to control virus infection.
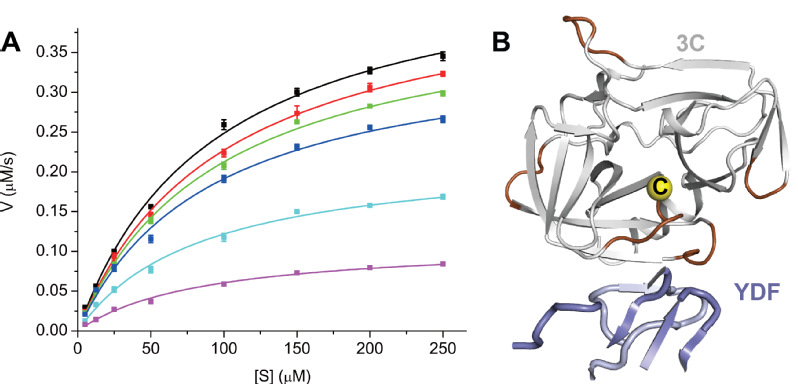
Figure 2. YDF allosterically inhibits the protease activity of 3C (A) Protease activity (in µM s-1) of HRV 3C was plotted against the substrate concentration (µM) in different molar ratios of YDF. YDF reduces the Vmax of 3C without affecting the apparent Km of HRV, indicating that it is a non-competitive inhibitor. (B) Molecular simulation indicates that YDF binding likely modulates the activity of 3C through allosterically affecting the catalytic cysteine (yellow sphere) and its flanking regions. The largest RMSF changes (dark brown) upon YDF binding are mapped onto the 3C structure
Previously, 3C from different enteroviruses has been found to specifically interact with the cloverleaf structure in the 5' noncoding region (NCR) of corresponding virus. Nevertheless, although disruption of the interaction therein has been correlated to RNA replication abolishment and viral viability loss in polio and EV71 viruses, the actual interface between 3C and viral genomic RNA that accounts for this interaction has not been determined. Here, the research team utilized hydrogen-deuterium exchange coupled mass spectrometry (HDX-MS) to determine which regions contributed to this interaction (Figure 3A), and further unraveled that the anti-viral effect of GGVV was conferred by abolishing interaction between 3C and viral genome (Figure 3B). This finding thus serves as a critical proof-of-concept that the RNA-binding site is indeed another promising drug target on 3C, which, given its sequence and structural conservation across different enteroviruses, may even hold potential for pan-enterovirus antiviral screening and design.
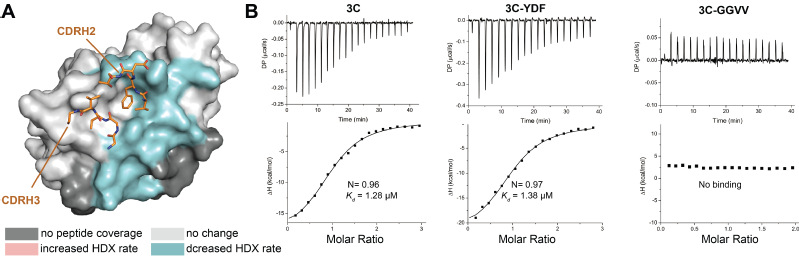
Figure 3. GGVV blocks the interaction between 3C and viral genomic RNA (A) The interface between 3C and viral genomic RNA overlaps with the epitope of GGVV. The structure of HRV 3C is shown as a surface representation, onto which the HDX difference heatmap for 3C in complex with genomic RNA compared to the apo form is mapped. Regions colored in cyan exhibited HDX rate decrease upon RNA binding, indicating their location at the interface between 3C and viral genomic RNA. (B) GGVV binding blocks interaction between 3C and the 5' noncoding regions of the viral genome
Taken together, this work identifies two novel sites of therapeutic vulnerability on an important antiviral target and provides an important framework for further rational inhibitor development against HRV and other enteroviruses.
Read the article: https://doi.org/10.1073/pnas.1918844117



