On November 3rd, 2020, a team led by SIAIS Distinguished Adjunct Professor Fan Guoping and Research Associate Professor Zhu Xianmin published new findings in the Springer Nature Group Journal Oncogene with the title of “Single-cell RNA sequencing reveals the heterogeneous tumor and immune cell populations in early-stage lung adenocarcinomas harboring EGFR mutations”. This work is based on the high-throughput single-cell RNA-seq analysis of over one hundred and twenty thousand single cells of lung adenocarcinoma (LUAD) and surrounding control cells. Using bioinformatics tools, the research team revealed heterogeneity of early-stage LUAD tumor cells and immune microenvironment, and discovered new drug targets for the treatment of early-stage LUAD.
Lung adenocarcinoma (LUAD) is the most common subtype of non-small cell lung cancer (NSCLC) and has high incidence in women, young patients and never smokers. EGFR mutation rates in LUAD are generally higher in Asian women. Understanding the inter-patient and intra-tumoral heterogeneity of LUAD is of great importance for precision medicine by refining the drug targets and developing new treatment strategies. However, previous single-cell RNA sequencing (scRNA-seq) analysis on NSCLC did not focus on a particular tumor type at a specific progression stage.
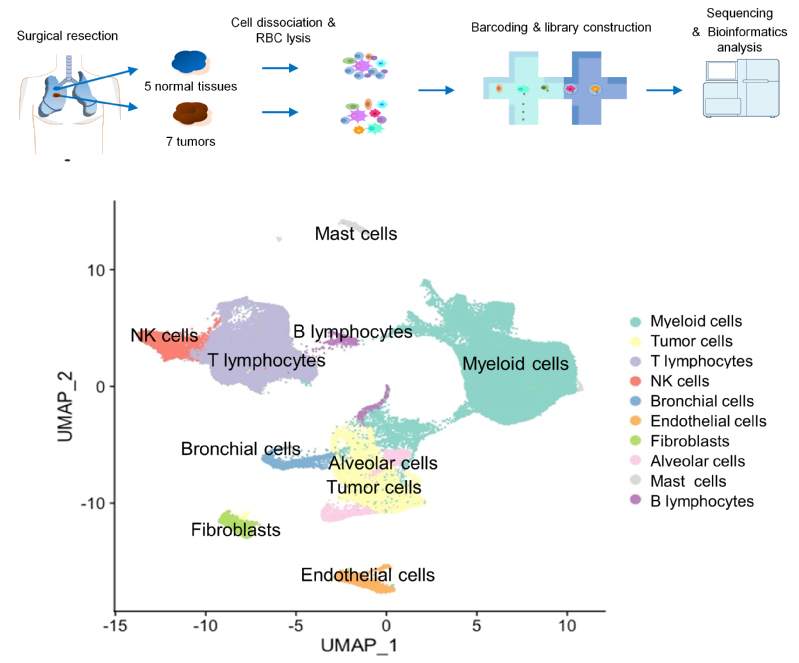
In this work, the researchers studied early-stage LUAD with EGFR mutations. To investigate the interactions between different cell types, they prepared single-cell suspensions of all the tumor and stromal cells from the resected tumor samples and their matched tumor-adjacent lung tissues. In total, the researchers collected 125,674 cells’ high quality transcriptomic data. From that, they reconstructed the tumor microenvironment (TME) that was composed of diverse cell types. In the TME, myeloid cells and T cells were the most abundant stromal cell types in the tumor samples and adjacent lung tissues. The tumor associated macrophages (TAMs) showed pro-tumoral functions in a M1/M2 mixed state. Tumor infiltrating T cells mainly displayed exhausted and regulatory T cell features, which was associated with the increase of CD1C+ dendritic cells and chemokines secreted by TAMs. The tumor cells were categorized into different subtypes based on their gene expression enrichment in distinct pathways such as hypoxia, glycolysis, cell metabolism, translation initiation, cell cycle and antigen presentation. When using the pseudotime trajectory to analyze the heterogeneity of tumor cells, the researchers identified ELF3 as one of the most upregulated genes in more advanced tumor cells. In response to secretion of inflammatory cytokines (e.g., IL1B) from the immune infiltrates, ELF3 in tumor cells was upregulated to trigger the activation of PI3K/Akt/NF-κB pathway and elevated expression of proliferation and anti-apoptosis genes.
Taken together, the researchers found substantial heterogeneity in tumor cells and surrounding stromal cells from early-stage LUAD harboring EGFR mutations, revealing that the interactions among tumor cells and different components in the TME would be potential new drug targets for LUAD treatment.
This work is led by Drs. Fan Guoping and Zhu Xianmin of ShanghaiTech University, as well as Dr. Zhang Peng from Shanghai Pulmonary Hospital. Postdoc fellow He Di of Memorial Sloan-Kettering Cancer Center in New York City, USA, is the first author of this paper.
Link:
https://www.nature.com/articles/s41388-020-01528-0



