On February 11, 2021, a research article entitled "Mediator structure and conformation change" was published in Molecular Cell. This report, based on a joint effort of the Lab of Structure Biochemistry at Shanghai Institute for Advanced Immunochemical Studies (SIAIS) and collaborators from Stanford describes, for the first time, the cryo-EM structure of the entire Mediator at a near-atomic resolution, and its conformation change before and after binding to RNA polymerase II (Pol II). This work provides an important basis for understanding the molecular mechanism of Mediator and represents a significant advance in basic research in the field of transcriptional regulation.
Transcription is one of the basic life activities of all prokaryotes and eukaryotes and is also an essential part of the central dogma. The main difference between eukaryotic transcription and prokaryotic transcription is that transcriptional activators bind directly to RNA polymerase, the core transcriptional machinery, in prokaryotes. In eukaryotes, transcriptional activators do not bind directly to Pol II but are mediated by a multiprotein complex called Mediator. In other words, in eukaryotes, the Mediator acts as a bridge between transcriptional activators and Pol II and it is required for the transcription of almost all eukaryotic genes.
Mediator was discovered in the lab of Prof. Roger D. Kornberg (2006 Nobel Prize Laureate in Chemistry). Mediator proved to contain many components previously identified in genetic screens for regulatory molecules in the lab of Richard A. Young among others. In 1999, the Kornberg Lab published a paper in Science, describing the overall shapes of yeast and murine Mediator complexes determined by negative-stain EM, demonstrating the generality of the Mediator complex. Since purification of the Mediator by the Kornberg Lab, many laboratories around the world have been trying to solve its structures. The yeast Mediator contains 21 subunits and can be divided into three modules termed Head, Middle and Tail. Due to its complex composition and flexibility, determining high-resolution structures of the entire Mediator has been a great challenge. In 2011 and 2012, two crystal structures of the Head module (of 4.3 Å and 3.4 Å, respectively) were reported in Nature. In 2017, a 4.4 Å cryo-EM map and a 3.4 Å crystal structure of the Head-Middle module were respectively reported in Nature. Although multiple high-resolution structures of several Mediator subcomplexes have been reported, high-resolution structures of the Tail module (400 kDa, a target of transcriptional activators) and the entire Mediator remained elusive, therefore limiting our understanding of how Mediator is involved in transcriptional regulation.
It is difficult to obtain high-resolution structures of yeast and mammalian Mediator complexes due to their conformational and compositional heterogeneity, so Dr. Zhang Heqiao (Research Assistant Professor of the Lab of Structure Biochemistry) took an unconventional approach. He chose to study the Mediator from Chaetomium thermophilum (a thermophilic fungus) whose proteins are of higher thermal stability and therefore more suitable for high-resolution structural analysis. First, he introduced an affinity tag at the C terminus of the Med6 subunit of the Head module via genetic manipulation. Then he succeeded in purifying the Mediator and Mediator-Pol II complexes from ~2 kg Chaetomium thermophilum cell pellet harvested from a 100-liter fermenter. By collaborating with Dr. Dong-hua Chen, a Senior Research Scientist in the Kornberg Lab at Stanford, he successfully determined the cryo-EM structures of the entire Mediator (6.6 Å) and its complex with Pol II (4.3 Å), in which the Head-Middle and Tail modules are of 4.2 Å and 3.6 Å respectively. The structure reveals the major activator-binding regions in Mediator. Additionally, by structural comparison, conformation changes of Mediator before and after Pol II binding were disclosed. After binding Pol II, the Tail module rotates, the Med1 subunit shifts slightly, and the gap at the Pol II-CTD (C-terminal domain) binding site closes. These conformation changes provide important clues for understanding the molecular mechanisms of Mediator in transcriptional regulation.
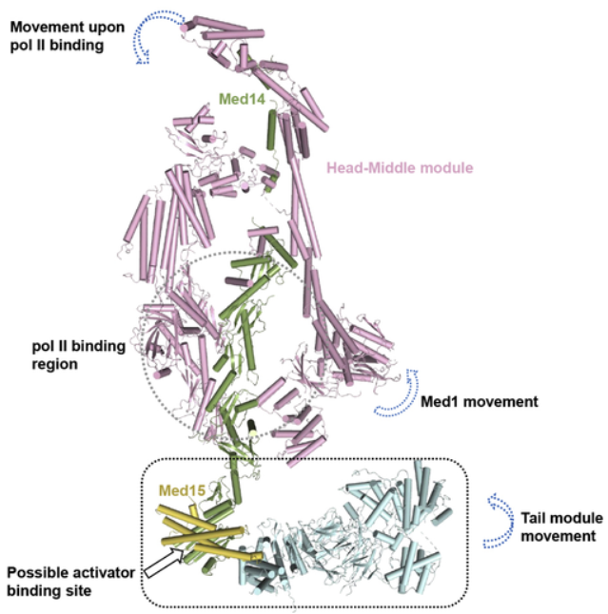
Figure 1. The cryo-EM structure of the entire Mediator at near-atomic resolution, and its conformation change before and after binding to Pol II
Notably, during the later stages, this project encountered challenges associated with the outbreak and spread of COVID-19, but the collaborators from China and the United States overcame all difficulties through close cooperation and timely communication, and therefore ensured the smooth completion of the project. Dr. Zhang Heqiao was the first author of this paper. Prof. Roger D. Kornberg (lead contact) and Dr. Nie Yan (Co-PI of the Lab of Structure Biochemistry) were co-corresponding authors, and ShanghaiTech University was the first affiliation. This project was supported by a grant from ShanghaiTech University.
Link to the article: https://doi.org/10.1016/j.molcel.2021.01.022

Figure 2. Group photo of all the authors (from left, Wang Lin, Wang Chunnian, Zhang Heqiao, Roger D. Kornberg, Nie Yan, Yuan Chao, Wang Yannan)



