| | 白芳 博士 |
Scientific Director of Biomedical Big Data Platform
课题组长(PI) 助理教授 所在学院 免疫化学研究所 生命科学与技术学院
研究方向 药物发现与设计;理论生物物理 联系方式 baifang@shanghaitech.edu.cn

教育经历
2009年毕业于大连理工大学,获化学工程与工艺与英语双学士学位; 2014年毕业于大连理工大学(上海药物所联合培养),获生物医学工程博士学位

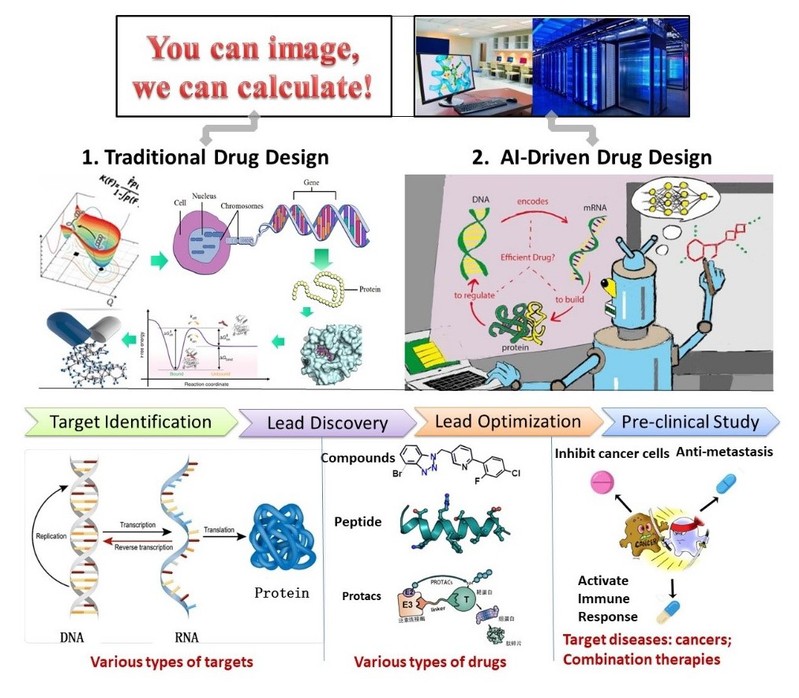
工作经历 2014.8-2019.5 美国莱斯大学,博士后; 2019.5-2019.9 美国德克萨斯大学休斯顿健康科学中心,研究助理教授; 2019.10-至今上海科技大学,任助理教授、研究员、课题组长  主要研究内容 1. 计算机辅助药物设计方法及高性能算法的开发。结合人工智能算法与传统的计算机辅助药物设计理论,发展诸如基于片段的药物分子设计方法、PROTACs设计方法、大尺度的蛋白质构象采样方法等。 2. 抗癌药物的发现与设计。利用课题组自有的计算方法与第三方计算方法,特别靶向肿瘤转移与免疫逃逸开展药物设计与优化研究。
 代表性论文 A. Amir1, F. Bai1,Y. Sohn, L. Song, S. Tamir, H. Marjault, G. Mayer, O. Karmi, P. Jennings, R. Mittlerf, J.N. Onuchic, A. Friedler, and R. Nechushtai. The anti-apoptotic proteins NAF-1 and iASPP interact to drive apoptosis in cancer cells. Chem. Sci. 2019, 10: 665-673(1Co-first Author). F. Bai, X. Pi, P. L, P. Zhou, H. Yang, X. Wang, M. Li, Z. Gao, H. Jiang. A Statistical Thermodynamic Model for Ligands Interacting With Ion Channels: Theoretical Model and Experimental Validation of the KCNQ2 Channel. Front. Pharmacol. 2018, 9:150. F. Bai, K. Liu, H. Li, J. Wang, J. Zhu, P. Hao, L. Zhu, S. Zhang, L. Shan, W. Ma, W. Zhang, H. Li, A.M. Bode &Z. Dong. Veratraimine modulates AP-1-depdendent gene transcription by directly binding to programmable DNA. Nucleic Acids Res. 2017, 46:546-557. F. Bai, F. Morcos, RR. Cheng, H. Jiang and J.N. Onuchic. Elucidating the druggable interface of protein-protein interactions using fragment-docking and coevolutionary analysis. Proc. Natl. Acad. Sci. USA. 2016, 113: E8051–E8058. F. Bai, F. Morcos, Y.S. Sohn, Darash-Yahana M., C.O. Rezende, C.H. Lipper, M.L. Paddock, L. Song, Y. Luo, S.H. Holt, S. Tamir, E.A. Theodorakis, P.A. Jennings, J.N. Onuchic, R. Mittler, and R. Nechushtai. The Fe-S cluster-containing NEET proteins mitoNEET and NAF-1 as chemotherapeutic targets in breast cancer. Proc. Natl. Acad. Sci. USA. 2015, 12: 3698-3703. F.Bai, S. Liao, J. Gu, H. Jiang, X. Wang, H. Li. An accurate metalloprotein-specific scoring function and molecular docking program devised by a dynamic sampling and iteration optimization strategy. J. Chem. Inf. Model. 2015, 55: 833-847. F.Bai, Y.Xu, J.Chen, Q.Liu, J.Gu, X.Wang, J.Ma, H. Li, J.N. Onuchic, and H.Jiang. Free energy landscape for the binding process of Huperzine A to acetylcholinesterase.Proc. Natl. Acad. Sci. USA. 2013, 110:4273-4278. F.Bai, H. Liu, L. Ton, W. Zhou, L. Liu, Z. Zhao, X. Liu, H. Jiang, X. Wang, H. Xie, and H. Li Discovery of novel selective inhibitors for EGFR-T790M/L858R. Bioorg. Med. Chem. Lett. 2012, 22:1365-1370. F.Bai, X. Liu, J. Li, H. Zhang, H. Jiang, X. Wang, and H. Li. Bioactive Conformational Generation of Small Molecules: A Comparative Analysis between Force-Field and Multiple Empirical Criteria Based Methods.BMC Bioinformatics 2010, 11:545. C. Lipper, J. Stofleth, F. Bai, Y. Sohn, S. Roy, R. Mittler, R. Nechushtai, J.N. Onuchic, P. Jennings. Redox-dependent gating of VDAC by mitoNEET. Proc. Natl. Acad. Sci. USA. 2019, 116: 19924-19929. Z. Tan, L. Xiao, M. Tang, F. Bai, J. Li, L. Li, F. Shi, N. Li, Y. Li, Q. Du, J. Lu, X. Weng, W. Yi, H. Zhang, J. Zhou, Q. Gao, J.N. Onuchic, A.M. Bode, X. Luo, Y. Cao. Targeting CPT1A-mediated fatty acid oxidation sensitizes nasopharyngeal carcinoma to radiation therapy. Theranostics 2018; 8:2329-2347. K. Yim, T. Prince, S. Qu, F. Bai, P. Jennings, J.N. Onuchic, E. Theodorakis and L. Neckers Gambogic acid identifies an isoform-specific druggable pocket in the middle domain of Hsp90β.Proc. Natl. Acad. Sci. USA. 2016, 113:E4801-E4809. M. Yahana, Y. Pozniak, M. Lu, Y. Sohn, O. Karmi, S. Tamir, F. Bai, L. Song, P. Jennings, E. Pikarsky, T. Geiger, J.N. Onuchic, R. Mittler and R. Nechushtai. Breast cancer tumorigenicity is dependent on high expression levels of NAF-1 and the lability of its Fe-S clusters. Proc. Natl. Acad. Sci. USA. 2016,113:10890-10895 Wang, Y. Jiang, J. Ma, H. Wu, D. Wacker, V. Katritch, G. Han, W., Liu W., Huang X-P., E. Vardy, J. McCorvy, X. Gao, X.E. Zhou, K. Melcher, C. Zhang, F.Bai, H. Yang, L. Yang, H. Jiang, B.L. Roth, V. Cherezov, RC. Stevens, and X.E. Xu. Structural Basis for Molecular Recognition at Serotonin Receptors. Science, 2013, 340:610-614.
|
|
|
|
